News
Computer Modeling Using SDSC Software Illustrates Plastic-Degrading Fungus
Published August 18, 2025
By Jack Imel and Kimberly Mann Bruch
Aspergillus oryzae, also known as koji mold, has been used to produce staple foods like soy sauce and miso for thousands of years. While this fungus has solved the age-old problem of satisfying our taste buds, it may also hold the key to solving a distinctly modern dilemma: global plastic pollution.
An international research team from the U.S. and Italy used computer modeling equipped with software developed at the San Diego Supercomputer Center (SDSC) at University of California San Diego’s School of Computing, Information and Data Sciences and Michigan State University (MSU) to study how cutinase, an enzyme found in koji mold, breaks down plastics at a molecular level. SDSC Associate Research Scientist Andreas Goetz was a key player in the work. Goetz said that the SDSC software helped power the study aimed at better understanding biotechnological approaches to break down plastics.
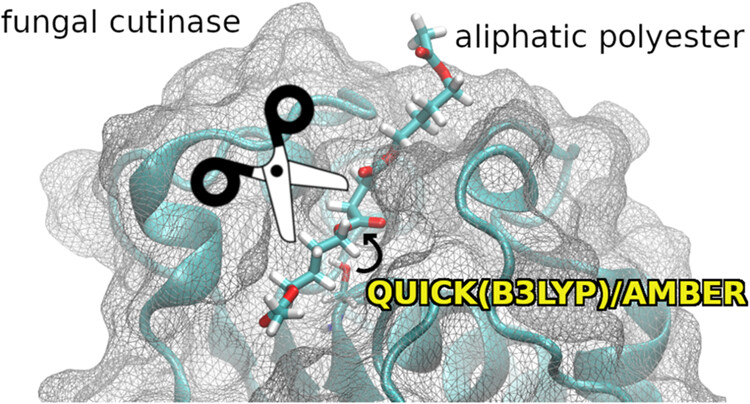
“This topic of using natural ways of degrading plastics has been studied for more than 50 years, but unlike past approaches that focused on engineering bacterial enzymes to degrade polyethylene terephthalate (PET) more effectively, our work targeted fungal cutinase enzymes degrading polybutylene succinate (PBS),” Goetz said. “We used software developed at SDSC and MSU called QUICK and Amber, while the simulations were run on a machine at the Istituto Italiano di Tecnologia in Italy (IIT), so this was truly an international collaboration.”
Goetz said that the research was led by colleagues from IIT and MSU. The team consisted of Goetz at SDSC, Marco De Vivo and Pietro Vidossich of IIT, along with Kenneth Merz and Madushanka Manathunga of MSU.
“Enzymes are a class of biological molecules found in all living things that act as catalysts — accelerating chemical reactions involved in digestion, metabolism, cellular signalling, muscle contraction and more,” explained Merz, who is the Joseph Zichis Chair in Chemistry at MSU. “In the human body enzymes called amylase, protease and lipase are responsible for digesting food, facilitating the breakdown of carbohydrates, proteins and fats through a chemical reaction called hydrolysis. In this chemical reaction — the same one catalyzed by cutinase in our study — complex molecules are split into smaller components by the addition of a water molecule. So, one might think of the process of cutinase degrading PBS as ‘digesting’ the plastic.”
Using SDSC’s QUICK and Amber, the team was able to observe how cutinase attached to the plastic substrate and how it broke the plastic’s chemical bonds, on an atomic level. They found that the cutinase enzyme was highly selective, preferentially targeting ester bonds — the fundamental building blocks of polyesters like PBS and PET.
“The observed ‘selectivity’ of cutinase is owed to its specific active site architecture,” explained Vidossich, lead author and IIT research associate. “The active site of an enzyme is the region where it binds with the substrate, PBS in our case. It’s like two puzzle pieces — only the right pieces will fit together, and they have to do so in a specific orientation. Cutinase is like a very special, very complicated puzzle piece.”
Additionally, they observed the degradation of the PBS substrate occurs in two steps with varying energy requirements: acylation and deacylation. They found that acylation, the initial binding of the enzyme, had a higher energy barrier than deacylation, when a water molecule enters the active site of the enzyme and hydrolyzes the plastic bond. Essentially, acylation was shown to be the energy ‘bottleneck’ in the process, limiting the rate at which the plastic can be degraded.
“With these insights, we provide guiding principles to engineer more efficient cutinase enzymes for breaking down a variety of polyesters that ultimately may help dealing with plastic waste,” Merz said. “Thanks to funding from the U.S. National Science Foundation (NSF), we are able to continue to extend the capabilities and performance of the software that includes techniques ranging from quantum mechanics to machine learning to enable such studies.”
The study was published in the Journal of Chemical Information and Modeling.
SDSC is a pillar of the UC San Diego School of Computing, Information and Data Sciences (SCIDS), which combines the strengths of SDSC, a national leader in high-performance and data-intensive computing, and the Halicioğlu Data Science Institute, a pioneering interdisciplinary institute that advances data science and AI education and research.
Support for the study included the National Centre for HPC, Big Data and Quantum Computing (grant no. CN00000013) and NSF (grant nos. OAC-2209717 and OAC-2435622).

